Ad-Hoc Code running in KubeFlow#
If you want to run a piece of code from your Jupyter notebook or a python script in kubeflow pipeline environment, you should use @run_in_kubeflow decorator.
Suppose you have a function my_func, relying on a sub-routine create_dataframe in your script/notebook. Having this second function might appear pointless, but you will see why we introduce it is a bit.
[ ]:
import numpy as np
import pandas as pd
np.random.seed(0)
def create_dataframe(array):
return pd.DataFrame(array)
def my_func(x: float, y: float, z: str):
matrix = np.random.rand(x, y)
df = create_dataframe(matrix)
df.index.name = z
return df.sum().sum()
We can run this function locally with some arbitrary inputs
[ ]:
my_func(x=3, y=5, z='random')
Modifying function to run in Kubeflow#
To run this function in kubeflow, we need to modify it in three ways:
1. Move all the imports inside the function#
For kubefow to serialise a function correctly, it should be self-sufficient, which means all the packages it uses and all the sub-routines it runs should be inside the function
[ ]:
def my_func(x: float, y: float, z: str):
# imports
import numpy as np
import pandas as pd
np.random.seed(0)
# define subroutines
def create_dataframe(array):
return pd.DataFrame(array)
# main function body
matrix = np.random.rand(x, y)
df = create_dataframe(matrix)
df.index.name = z
return df.sum().sum()
Notice that the results of the function will be identical if we run it locally
[ ]:
my_func(x=3, y=5, z='random')
2. Make function accept one input: input_dict of type dict#
Instead of supplying all of the arguments separately, you need to group them in a dictionary and parse this dictionary inside your function. Again, this is done so that octaikube can supply your function to kubeflow.
NOTE input_dict is not an example name, it is the precise name you have to use as input to your function.
[ ]:
def my_func(input_dict: dict):
# imports
import numpy as np
import pandas as pd
np.random.seed(0)
# define subroutines
def create_dataframe(array):
return pd.DataFrame(array)
# parse input dictionary
x = input_dict['x']
y = input_dict['y']
z = input_dict['z']
# main function body
matrix = np.random.rand(x, y)
df = create_dataframe(matrix)
df.index.name = z
return df.sum().sum()
The inputs to the function can then be defined as
[ ]:
input_dict = {'x': 3, 'y': 5, 'z': 'random'}
And the function is called this way will still work just like before locally
[ ]:
my_func(input_dict)
3. OctaiKube @run_in_kubeflow decorator#
Lastly, we need to decorate our function with octaikube decorator so that it will actually run on a cluster, rather than in python/ipython runtime.
@run_in_kubeflow takes one positional argument, experiment_name (str) which appends the run in existing kubeflow experiment or creates a new one if experiment under given name doesn’t exist.
Environmental variables#
To run your function on the cluster using run_in_kubeflow decorator, you need KUBEFLOW_HOST environment variable to be set in your runtime. If you are not sure what is the address of your kubeflow host, ask your administrator.
If you are running your code inside a jupyter notebook, it will not have access to your session environment variables. So to add your variables to the python runtime, you need to create and .env file which should look something like
KUBEFLOW_HOST=<kubeflow_host_value>
FOO=BAR
You then need to use dotenv extension to load the variables into the runtime.
[ ]:
%load_ext dotenv
%dotenv
[ ]:
from octaikube.adhoc import run_in_kubeflow
from dotenv import load_dotenv
load_dotenv(dotenv_path='.')
@run_in_kubeflow(experiment_name='adhoc_guide')
def my_func(input_dict: dict):
# imports
import numpy as np
import pandas as pd
np.random.seed(0)
# define subroutines
def create_dataframe(array):
return pd.DataFrame(array)
# parse input dictionary
x = input_dict['x']
y = input_dict['y']
z = input_dict['z']
# main function body
matrix = np.random.rand(x, y)
df = create_dataframe(matrix)
df.index.name = z
return df.sum().sum()
Now if we define input_dict and run the function as before, octaikube will send it to run on a cluster.
[ ]:
input_dict = {'x': 3, 'y': 5, 'z': 'random'}
my_func(input_dict)
After that in your kubeflow dashboard you will see the experiment under the name you have provided
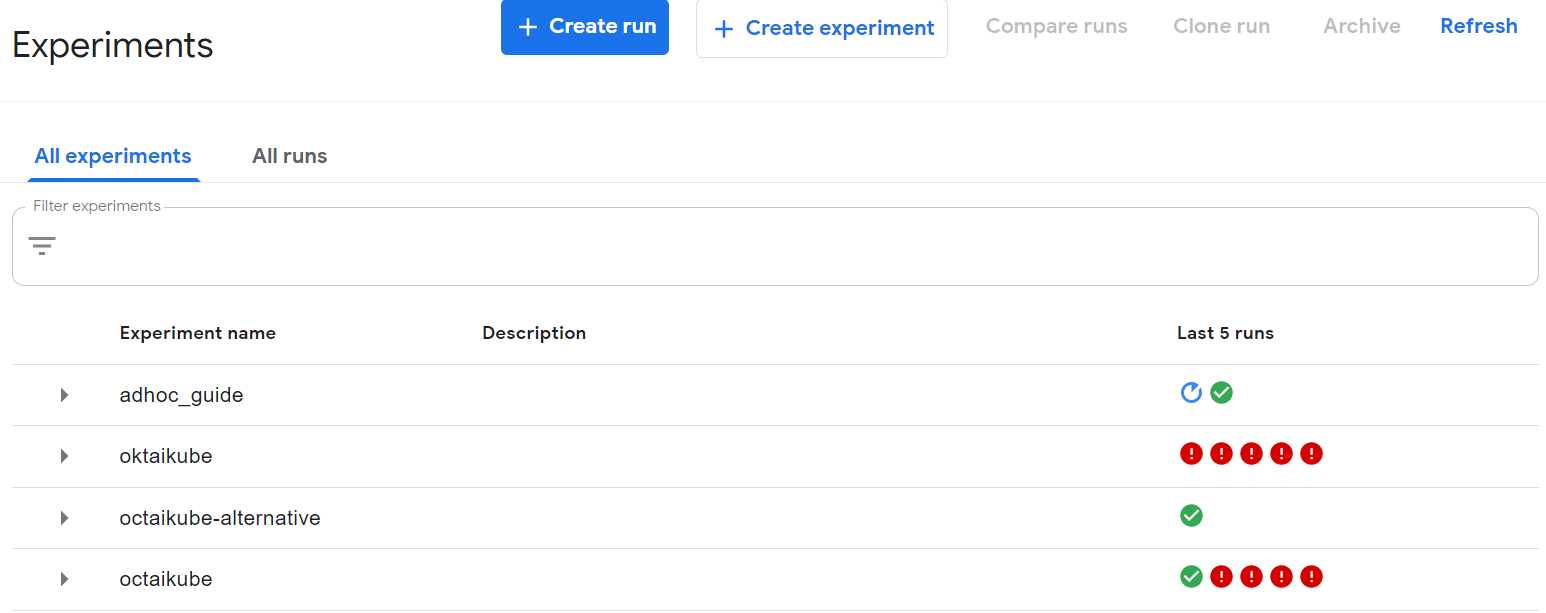
If you further navigate into the lates run in adhoc_guide experiment, you will see my_func pipeline component and clicking on it will show you information about it.
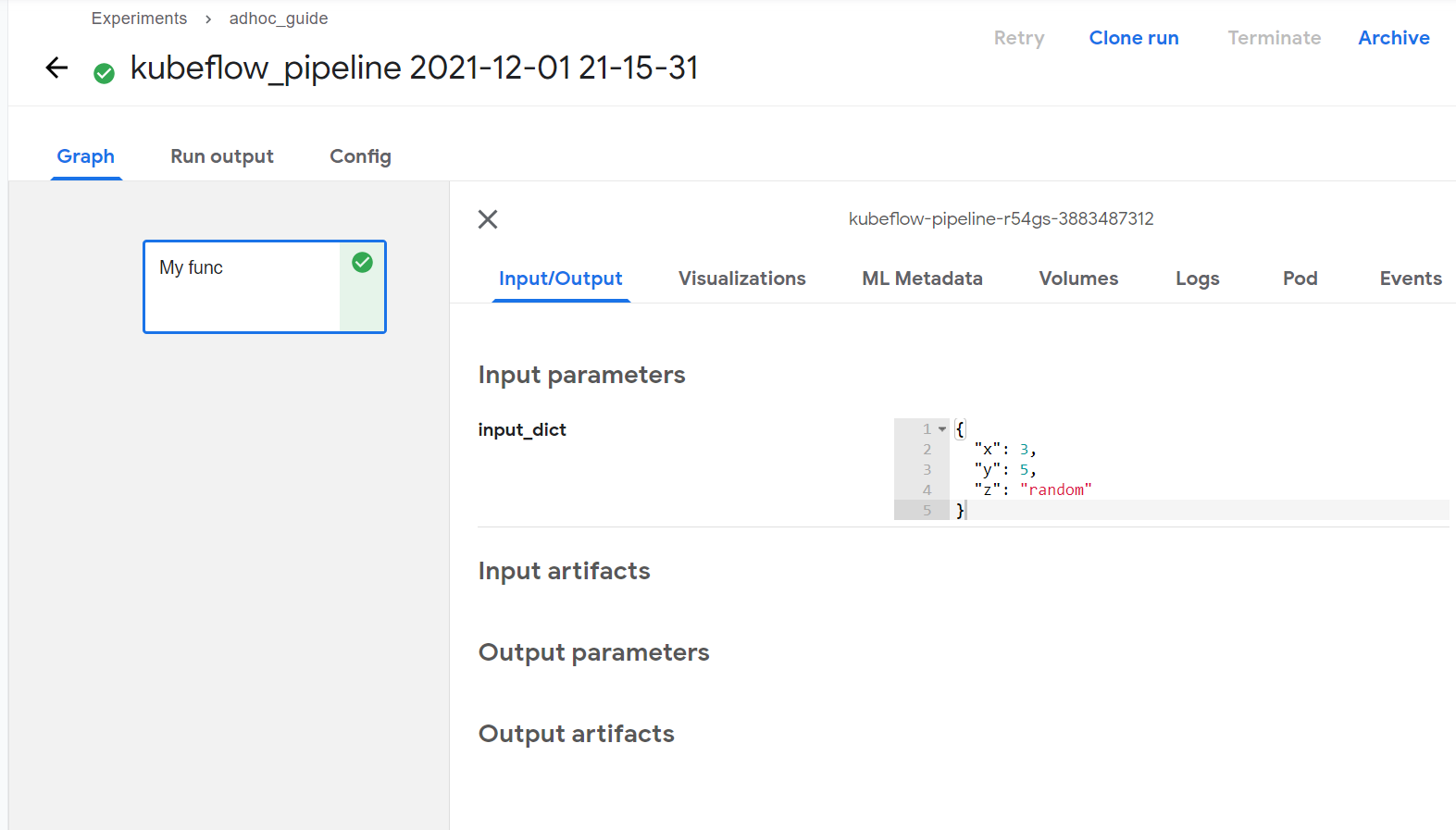
You can see the input_dict you specified in the ‘Input Parameters’ section.
NOTE ability to view/stream outputs back is being developed, for now, make sure all the results that your code is producing are saved on a network location, such as blob storage.
[ ]: